BisQue Project
BisQue Platform. The BisQue platform allows users to store/share, visualize, and analyze all of their image data (250+ image formats).
BisQue is a web-based platform designed to provide researchers with organizational and quantitative analysis tools for up to 5D image data. Users can extend BisQue by creating their own machine learning modules or building complex containerized workflows for preprocessing, inference, and postprocessing. Afterwards, they can visualize the results in the web browser using BisQue’s volumetric viewer. BisQue’s extensibility stems from two core concepts: flexible metadata facility and an open web-based architecture. Together these empower researchers to create, develop and share novel multimodal data analyses.
Production ML Modules
CellECT 2.0
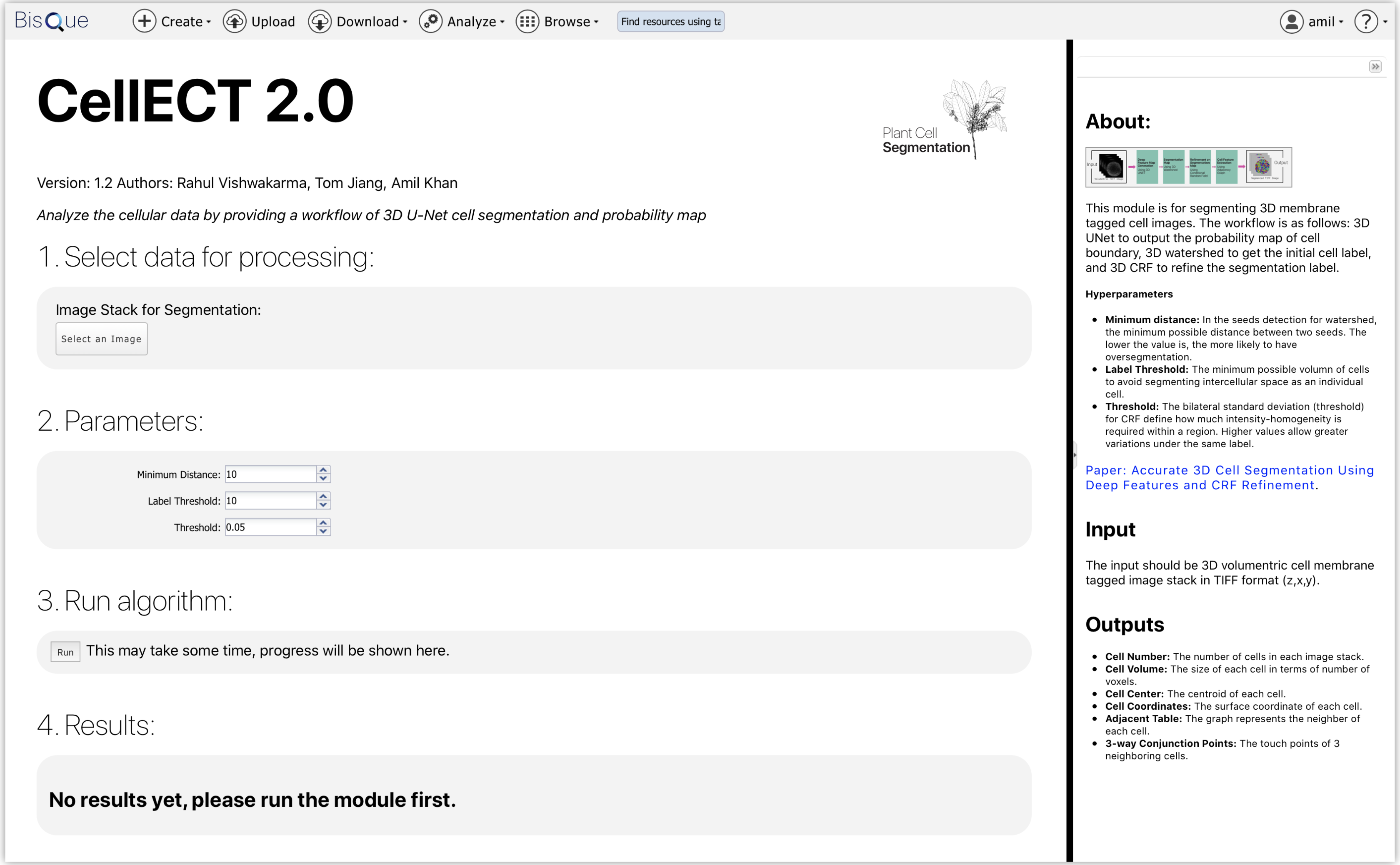
CellECT 2.0 Module. A deep learning cell segmentation module for plant cells.
This module is for segmenting 3D membrane tagged cell images. The workflow is as follows: 3D UNet to output the probability map of cell boundary, 3D watershed to get the initial cell label, and 3D CRF to refine the segmentation label.
NPH Prediction
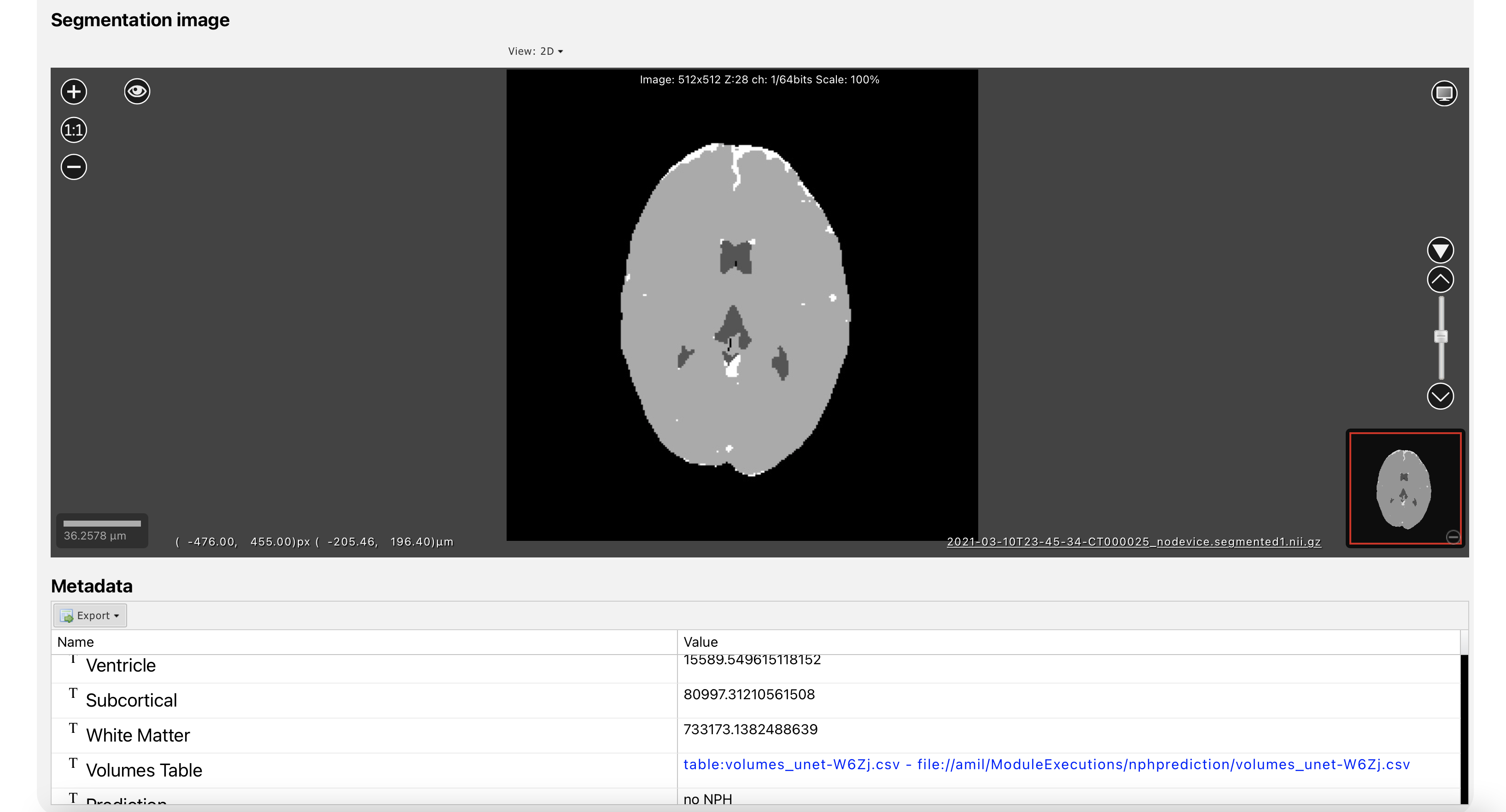
NPH Prediction Module Output. A deep learning for segmenting CT Scans and predicting possible NPH.
This module is for segmenting a CT scan of a brain into 3 parts: lateral ventricles, cerebral matter, and subarachnoid space using a 3D UNet. A Support Vector Machine is then used to determine if the subject has possible Normal Pressure Hydrocephalus (NPH) (Zhang, Khan, Manjunath, & others, 2021).
Composite Strength
This module implements a surrogate model that predicts effective yield strength of a composite given its 3-D microstructure. The model is developed for composites with strength contrast of 5: strength of hard phase \(\dfrac{s_2}{s_1} = 5\). The model works best on periodic 3-D microstructures.
Calibration of the model is done by finite element simulation data by multivariate polynomial regression between principal component scores of 2-point statistics (representing microstructure) and effective yield strength (property) (Latypov et al., 2019).